-
Antimicrobial resistance is a major clinical problem, with resistant strains of bacteria emerging at a rate that dramatically outpaces development of new drugs. Traditionally, studies on antibiotic resistance have focused on genetic changes that confer resistance, such as mutations that block the drug target. However, bacteria can also evade antibiotics through expression of transient resistance mechanisms, such as multi-drug efflux pumps. Expression of these genes can vary substantially from cell to cell within a population. We are interested in identified new genes involved in transient resistance, discovering mechanisms that generate heterogeneity in expression of antibiotic resistance genes, and linking these cell-to-cell differences resistance gene expression to mutation rate.
- N. M. V. Sampaio*, C. M. Blassick*, V. Andreani, J. B. Lugagne, M. J. Dunlop (*Equal contribution). "Dynamic Gene Expression and Growth Underlie Cell-to-Cell Heterogeneity in Escherichia coli Stress Response." Proceedings of the National Academy of Sciences, 2022.
- I. El Meouche, M. J. Dunlop. "Heterogeneity in Efflux Pump Expression Predisposes Antibiotic Resistant Cells to Mutation." Science, 2018.
-
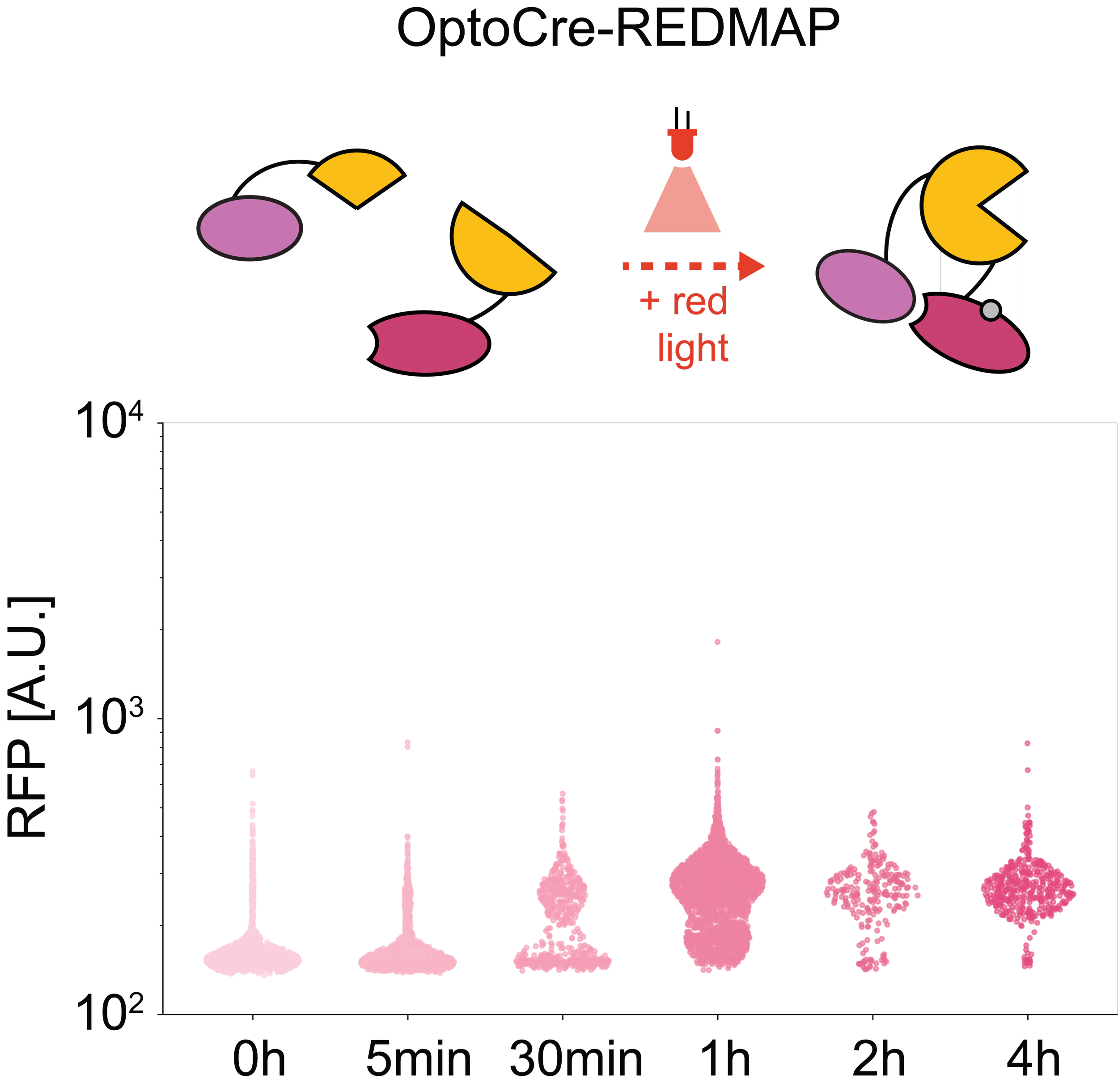
We are developing optogenetic tools and methods that work in bacteria. These approaches use light (typically from LEDs) to control gene expression. Because light can easily be turned on and off, and can be spatially patterned, it is offers an attractive alternative to the chemical inducers that are commonly used to control gene expression.
- F. Jafarbeglou, M. J. Dunlop. "Red Light Responsive Cre Recombinase for Bacterial Optogenetics." ACS Synthetic Biology, DOI: 10.1021/acssynbio.4c00388, 2024.
- M. B. Sheets, N. Tague, M. J. Dunlop. "An Optogenetic Toolkit for Light-Inducible Antibiotic Resistance." Nature Communications, https://doi.org/10.1038/s41467-023-36670-2, 2023.
-
Feedback allows biological systems to control gene expression precisely and reliably. However, experimental implementations of biomolecular control systems are still far from satisfying performance specifications typically achieved by electrical or mechanical control systems. We design genetic control systems and implement hybrid approaches that use computers to control gene expression (e.g. via light with optogenetics).
- C. M. Blassick, J. B. Lugagne, M. J. Dunlop. "Dynamic Heterogeneity in an E. coli Stress Response Regulon Mediates Gene Activation and Antimicrobial Peptide Tolerance." [bioRxiv preprint]
- J. B. Lugagne, C. M. Blassick, M. J. Dunlop. "Deep Model Predictive Control of Gene Expression in Thousands of Single Cells."
Nature Communications, https://doi.org/10.1038/s41467-024-46361-1, 2024.
-
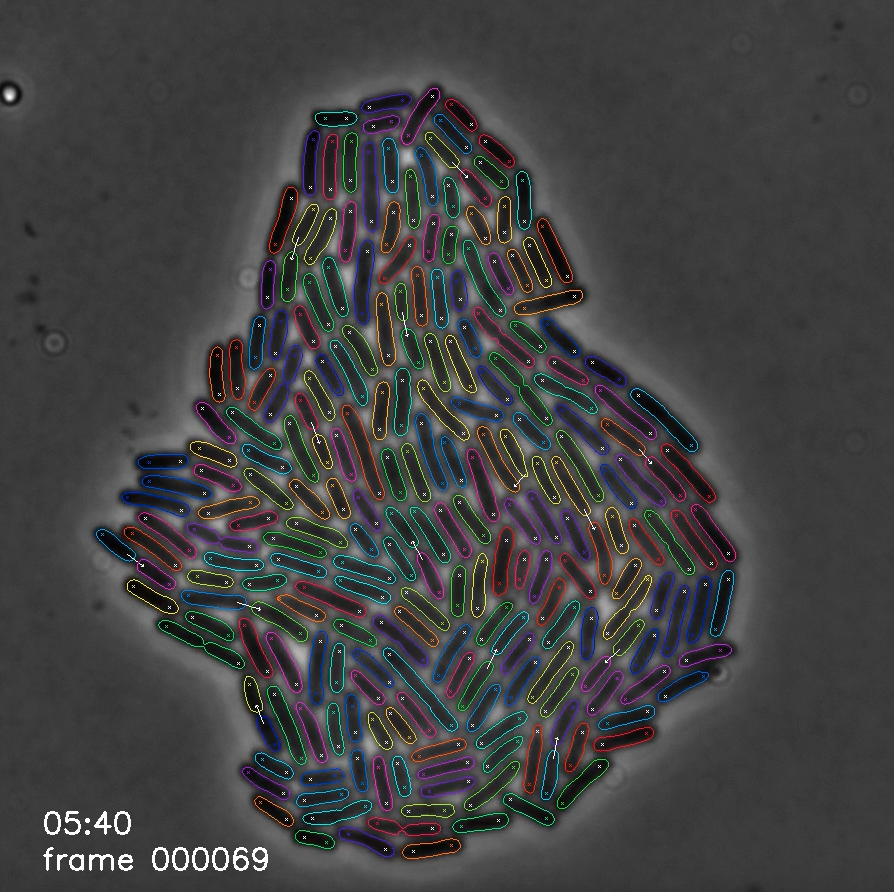
Image processing is a major bottleneck in the analysis pipeline for microscopy data. Traditionally, processing happens after full experiments are completed and software tools are specific to the cell type and imaging set up. In addition, this has historically been a cumbersome process requiring significant manual input and error correction. To address these challenges, we developed an image processing tool, Deep Learning for Time-Lapse Analysis (DeLTA), that uses two U-Net convolutional neural networks to rapidly and accurately segment and track cells within time-lapse images. We are developing new methods for reliably performing image analysis, including those that can be deployed in real-time.
- O. M. O’Connor, R. N. Alnahhas, J. B. Lugagne+, M. J. Dunlop+ (+Co-Corresponding). “DeLTA 2.0: A Deep Learning Pipeline for Quantifying Single-cell Spatial and emporal Dynamics.” PLOS Computational Biology, 2022.
- J. B. Lugagne, H. Lin, M. J. Dunlop. "DeLTA: Automated Cell Segmentation, Tracking, and Lineage Reconstruction using Deep Learning." PLOS Computational Biology, 2020.
-
Engineered microbes can produce valuable chemicals, however production strains often require extensive optimization. Furthermore, typical approaches for quantifying yield are derived from bulk cultures, obscuring potential cell-to-cell differences in production, and prohibiting selection methods that rely on measurements of a single cell’s phenotype, such as directed evolution. We develop new methods to directly visualize production in metabolically engineered cells.
- N. Tague*, H. Lin*, J. B. Lugagne, O. M. O’Connor, D. Burman, W. W. Wong, J.-X. Cheng, M. J. Dunlop (*Equal contribution). "Longitudinal Single‐Cell Imaging of Engineered Strains with Stimulated Raman Scattering to Characterize Heterogeneity in Fatty Acid Production." Advanced Science, 2023.
- T. Wang, M. J. Dunlop. "Controlling and Exploiting Cell-to-Cell Variation in Metabolic Engineering." Current Opinion in Biotechnology, 2018.